Expression-based machine learning models for predicting plant tissue identity
Abstract
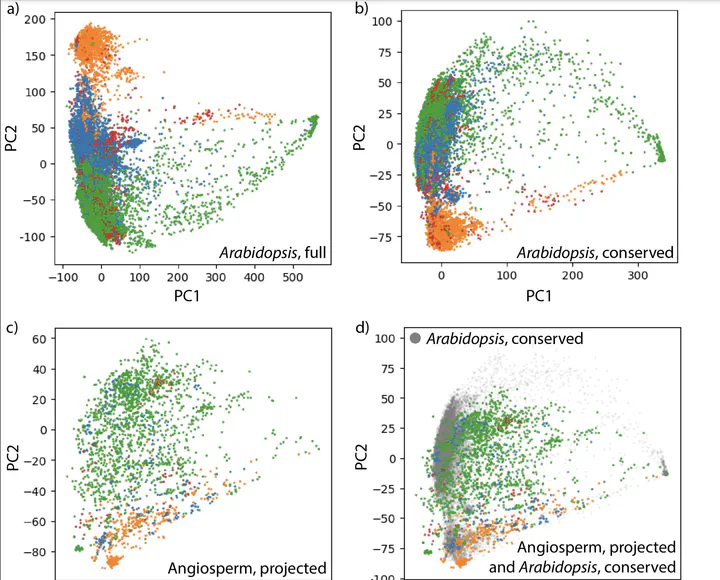
The selection of Arabidopsis as a model organism played a pivotal role in advancing genomic science, firmly establishing the cornerstone of today ‘s plant molecular biology. Competing frameworks to select an agricultural- or ecological-based model species, or to decentralize plant science and study a multitude of diverse species, were selected against in favor of building core knowledge in a species that would facilitate genome-enabled research that could assumedly be transferred to other plants. Here, we examine the ability of models based on Arabidopsis gene expression data to predict tissue identity in other flowering plant species. Comparing different machine learning algorithms, models trained and tested on Arabidopsis data achieved near perfect precision and recall values using the K-Nearest Neighbor method, whereas when tissue identity is predicted across the flowering plants using models trained on Arabidopsis data, precision values range from 0.69 to 0.74 and recall from 0.54 to 0.64, depending on the algorithm used. Below-ground tissue is more predictable than other tissue types, and the ability to predict tissue identity is not correlated with phylogenetic distance from Arabidopsis. This suggests that gene expression signatures rather than marker genes are more valuable to create models for tissue and cell type prediction in plants. Our data-driven results highlight that, in hindsight, the assertion that knowledge from Arabidopsis is translatable to other plants is not always true. Considering the current landscape of abundant sequencing data and computational resources, it may be prudent to reevaluate the scientific emphasis on Arabidopsis and to prioritize the exploration of plant diversity.
Type
Publication
bioRxiv